Note
Go to the end to download the full example code or to run this example in your browser via Binder
n. spider plot
This file shows the usage of spider_plot() function.
# sphinx_gallery_thumbnail_number = -2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from easy_mpl import spider_plot
from easy_mpl.utils import version_info, create_subplots
version_info()
{'easy_mpl': '0.21.4', 'matplotlib': '3.8.0', 'numpy': '1.26.1', 'pandas': '2.1.1', 'scipy': '1.11.3'}
specifying labels
# specifying tick size
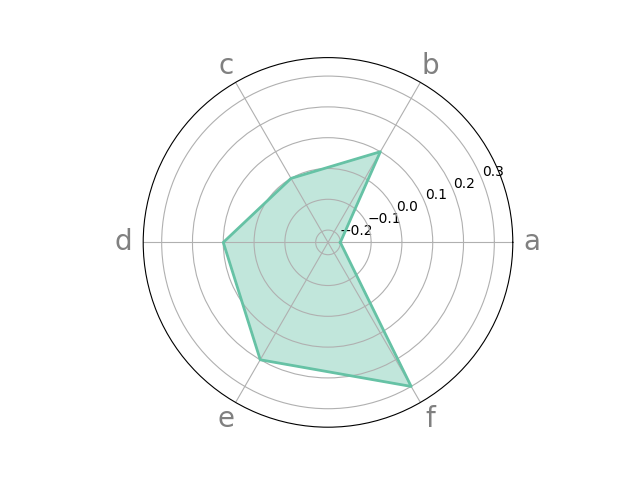
# specifying colors on our own
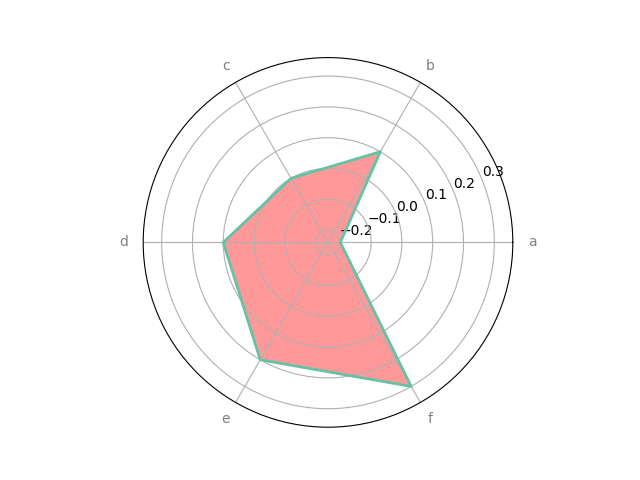
# specifying outline color
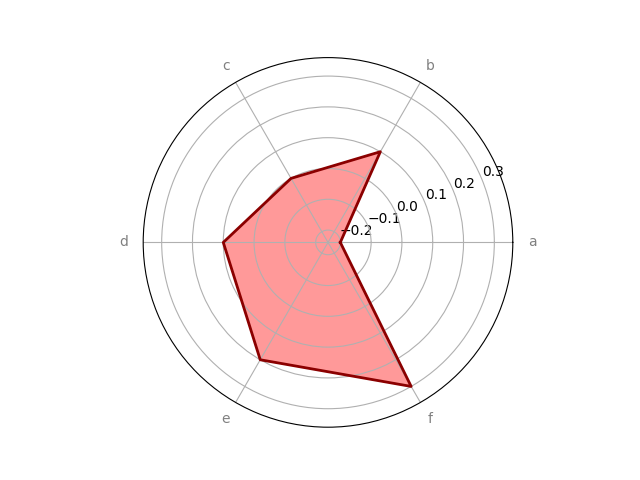
# we can also specify cmap in place of color
Using dataframe
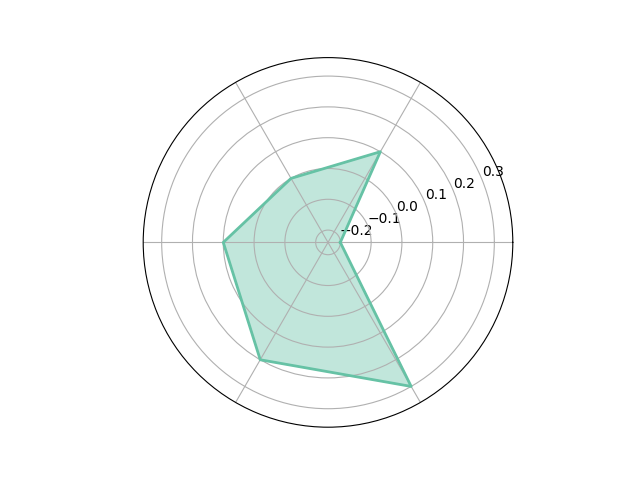
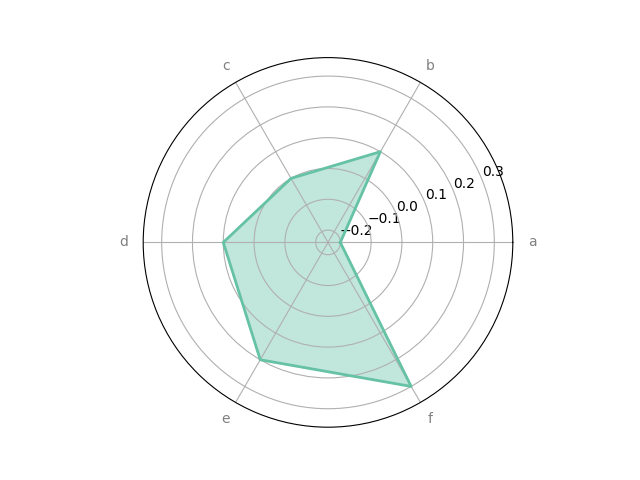
df = pd.DataFrame.from_dict(
{'summer': {'a': -0.2, 'b': 0.1, 'c': 0.0, 'd': 0.1, 'e': 0.2, 'f': 0.3},
'winter': {'a': -0.3, 'b': 0.1, 'c': 0.0, 'd': 0.2, 'e': 0.15,'f': 0.25}})
_ = spider_plot(df, xtick_kws={'size': 20})
# #%%
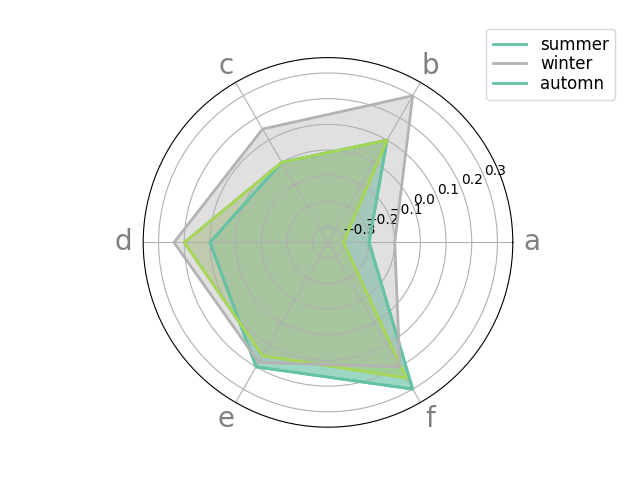
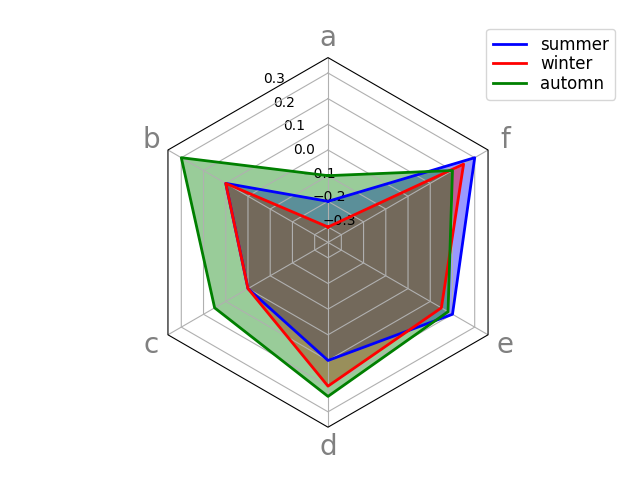
df = pd.DataFrame.from_dict(
{'summer': {'a': -0.2, 'b': 0.1, 'c': 0.0, 'd': 0.1, 'e': 0.2, 'f': 0.3},
'winter': {'a': -0.3, 'b': 0.1, 'c': 0.0, 'd': 0.2, 'e': 0.15, 'f': 0.25},
'automn': {'a': -0.1, 'b': 0.3, 'c': 0.15, 'd': 0.24, 'e': 0.18, 'f': 0.2}})
_ = spider_plot(df, xtick_kws={'size': 20})
# #%%
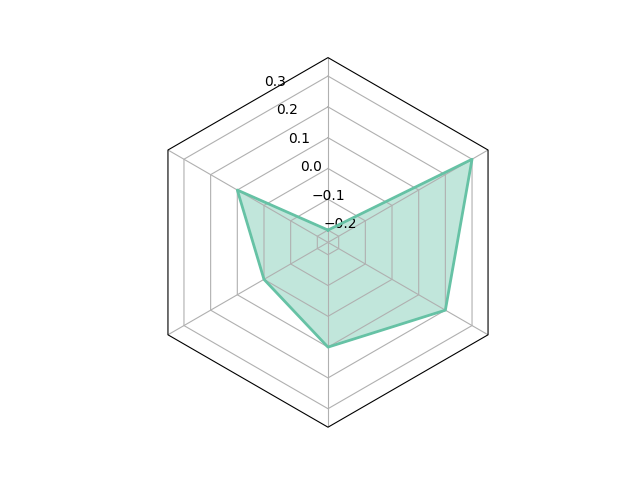
# use polygon frame
_ = spider_plot(data=values, frame="polygon")
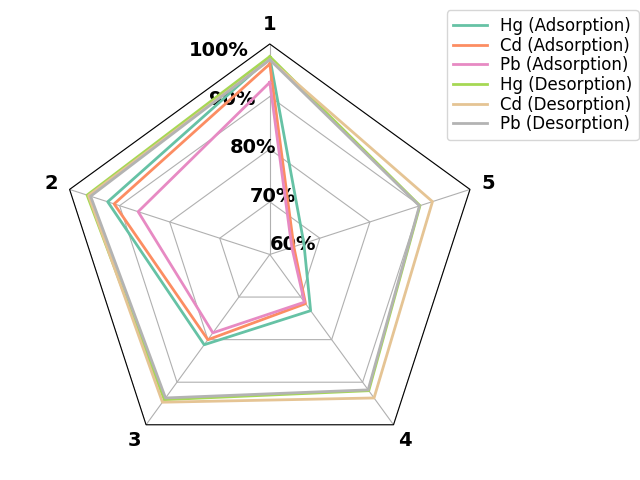
postprocessing of axes
df = pd.DataFrame.from_dict(
{
'Hg (Adsorption)': {'1': 97.4, '2': 92.38, '3': 81.2, '4': 73.2, '5': 66.81},
'Cd (Adsorption)': {'1': 96.2, '2': 91.1, '3': 80.02, '4': 71.55, '5': 64.8},
'Pb (Adsorption)': {'1': 92.7, '2': 86.3, '3': 78.4, '4': 71.2, '5': 64.4},
'Hg (Desorption)': {'1': 97.6, '2': 96.5, '3': 94.1, '4': 91.99, '5': 90.0},
'Cd (Desorption)': {'1': 97.0, '2': 96.2, '3': 94.7, '4': 93.7, '5': 92.5},
'Pb (Desorption)': {'1': 97.0, '2': 95.8, '3': 93.7, '4': 91.8, '5': 89.9}
})
_ = spider_plot(df, frame="polygon",
fill_kws = {"alpha": 0.0},
xtick_kws={'size': 14, "weight": "bold", "color": "black"},
show=False,
leg_kws = {'bbox_to_anchor': (0.90, 1.1)}
)
plt.gca().set_rmax(100.0)
plt.gca().set_rmin(60.0)
plt.gca().set_rgrids((60.0, 70.0, 80.0, 90.0, 100.0),
("60%", "70%", "80%", "90%", "100%"),
fontsize=14, weight="bold", color="black"),
plt.tight_layout()
plt.show()
matplotlib example
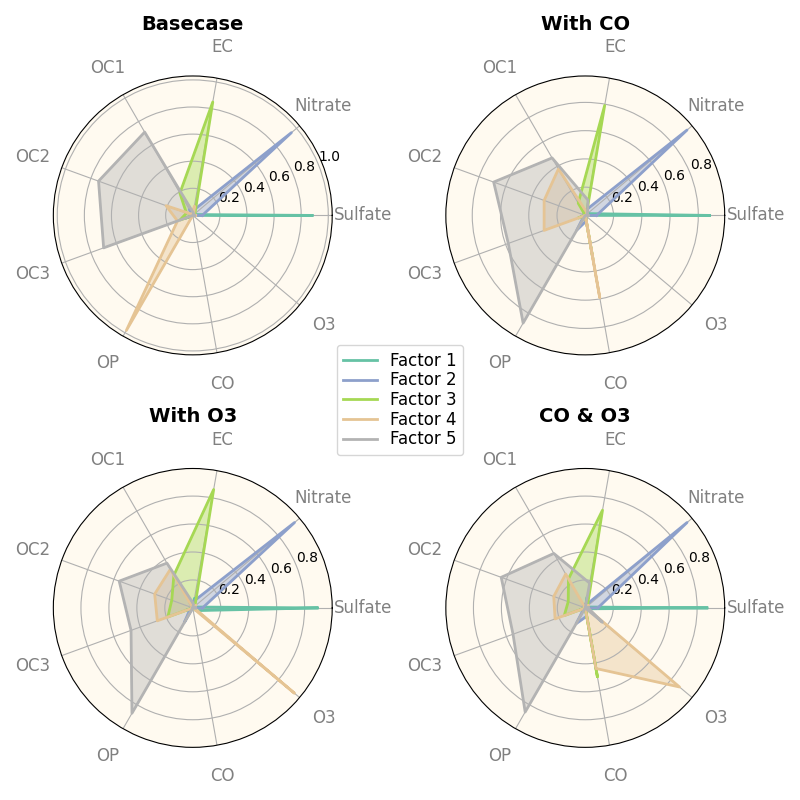
data = [
['Sulfate', 'Nitrate', 'EC', 'OC1', 'OC2', 'OC3', 'OP', 'CO', 'O3'],
('Basecase', [
[0.88, 0.01, 0.03, 0.03, 0.00, 0.06, 0.01, 0.00, 0.00],
[0.07, 0.95, 0.04, 0.05, 0.00, 0.02, 0.01, 0.00, 0.00],
[0.01, 0.02, 0.85, 0.19, 0.05, 0.10, 0.00, 0.00, 0.00],
[0.02, 0.01, 0.07, 0.01, 0.21, 0.12, 0.98, 0.00, 0.00],
[0.01, 0.01, 0.02, 0.71, 0.74, 0.70, 0.00, 0.00, 0.00]]),
('With CO', [
[0.88, 0.02, 0.02, 0.02, 0.00, 0.05, 0.00, 0.05, 0.00],
[0.08, 0.94, 0.04, 0.02, 0.00, 0.01, 0.12, 0.04, 0.00],
[0.01, 0.01, 0.79, 0.10, 0.00, 0.05, 0.00, 0.31, 0.00],
[0.00, 0.02, 0.03, 0.38, 0.31, 0.31, 0.00, 0.59, 0.00],
[0.02, 0.02, 0.11, 0.47, 0.69, 0.58, 0.88, 0.00, 0.00]]),
('With O3', [
[0.89, 0.01, 0.07, 0.00, 0.00, 0.05, 0.00, 0.00, 0.03],
[0.07, 0.95, 0.05, 0.04, 0.00, 0.02, 0.12, 0.00, 0.00],
[0.01, 0.02, 0.86, 0.27, 0.16, 0.19, 0.00, 0.00, 0.00],
[0.01, 0.03, 0.00, 0.32, 0.29, 0.27, 0.00, 0.00, 0.95],
[0.02, 0.00, 0.03, 0.37, 0.56, 0.47, 0.87, 0.00, 0.00]]),
('CO & O3', [
[0.87, 0.01, 0.08, 0.00, 0.00, 0.04, 0.00, 0.00, 0.01],
[0.09, 0.95, 0.02, 0.03, 0.00, 0.01, 0.13, 0.06, 0.00],
[0.01, 0.02, 0.71, 0.24, 0.13, 0.16, 0.00, 0.50, 0.00],
[0.01, 0.03, 0.00, 0.28, 0.24, 0.23, 0.00, 0.44, 0.88],
[0.02, 0.00, 0.18, 0.45, 0.64, 0.55, 0.86, 0.00, 0.16]])
]
fig, axes = create_subplots(4, subplot_kw= dict(projection='polar'),
figsize=(8,8))
spoke_labels = data.pop(0)
labels = ('Factor 1', 'Factor 2', 'Factor 3', 'Factor 4', 'Factor 5')
for ax, (title, col) in zip(axes.flat, data):
d = np.array(col).transpose()
ax.set_title(title, fontdict={'fontsize': 14, 'fontweight':'bold'})
_ = spider_plot(d, show=False, ax=ax,
tick_labels=spoke_labels, xtick_kws=dict(size=12)
)
ax.tick_params(axis='x', which='major', pad=12)
ax.set_facecolor('floralwhite')
fig.legend(labels, loc='center', labelspacing=0.1, fontsize='large')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 5.747 seconds)